Structural dynamics of the SARS-CoV-2 frameshift-stimulatory pseudoknot reveal topologically distinct conformers | bioRxiv
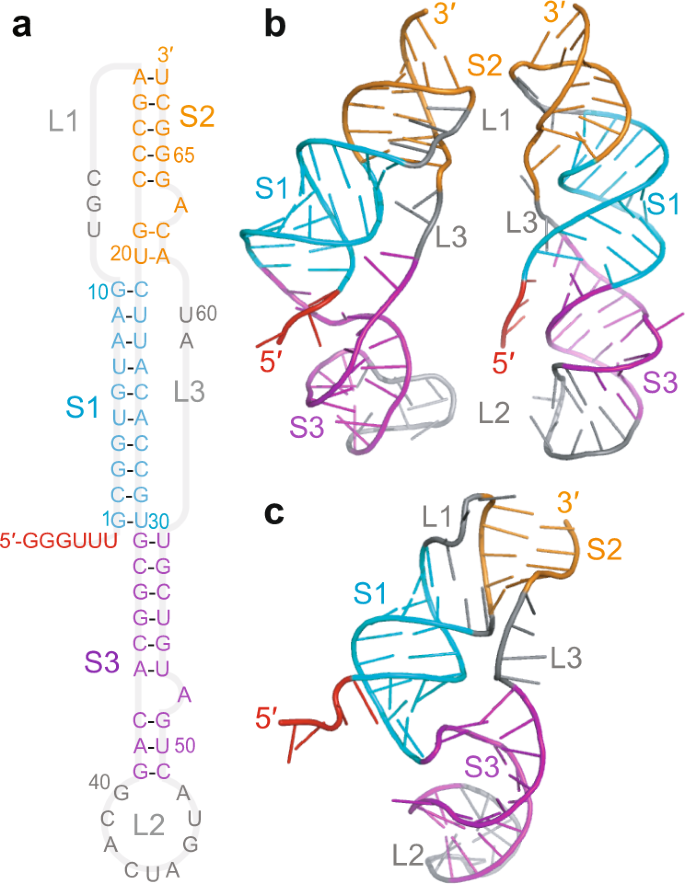
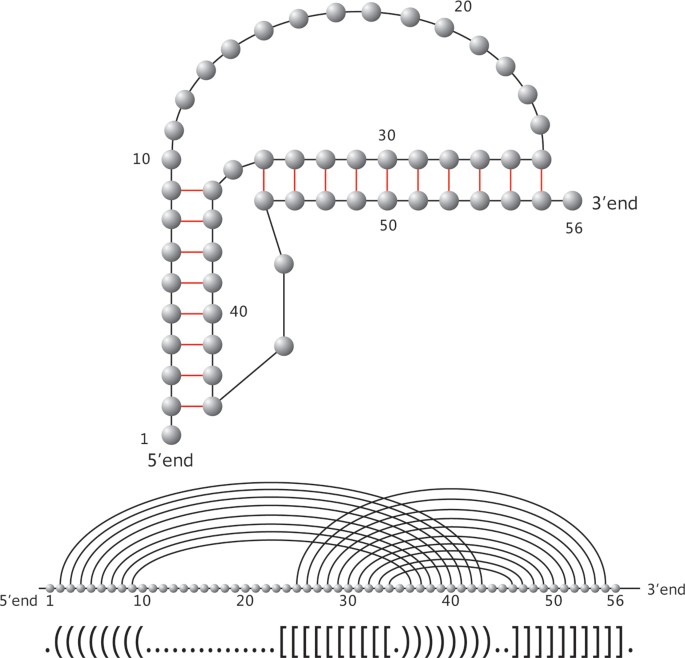
Structural dynamics of single SARS-CoV-2 pseudoknot molecules reveal topologically distinct conformers | Nature Communications
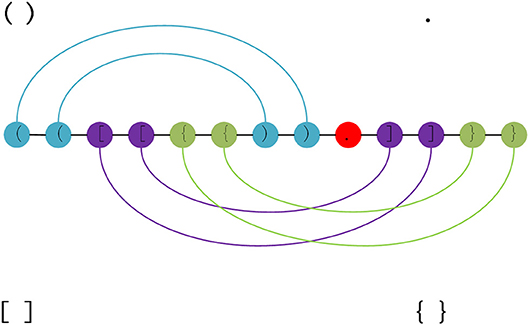
Frontiers | DMfold: A Novel Method to Predict RNA Secondary Structure With Pseudoknots Based on Deep Learning and Improved Base Pair Maximization Principle
Mapping in Solution Shows the Peach Latent Mosaic Viroid To Possess a New Pseudoknot in a Complex, Branched Secondary Structure
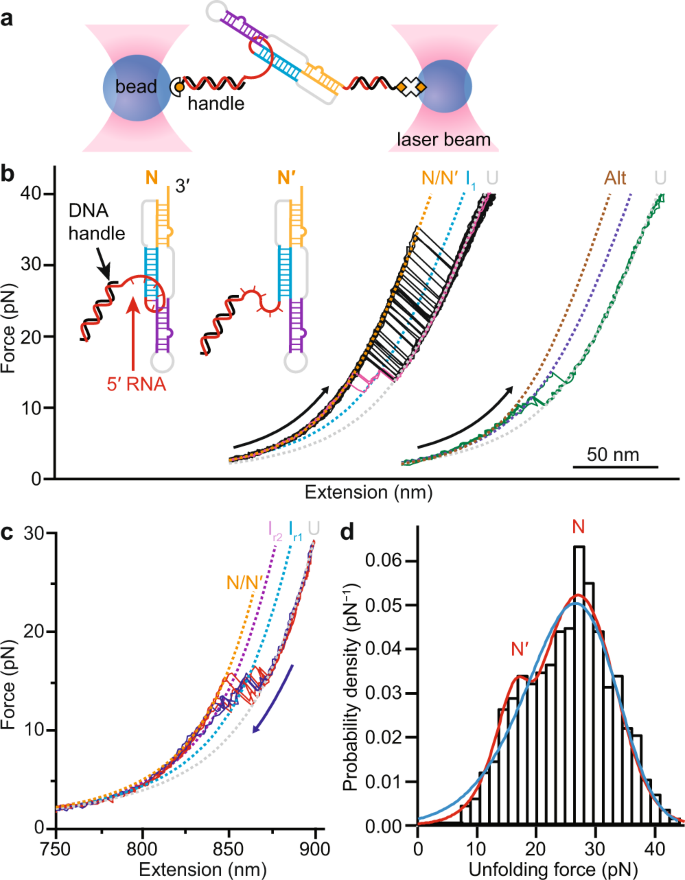
Structural dynamics of single SARS-CoV-2 pseudoknot molecules reveal topologically distinct conformers | Nature Communications
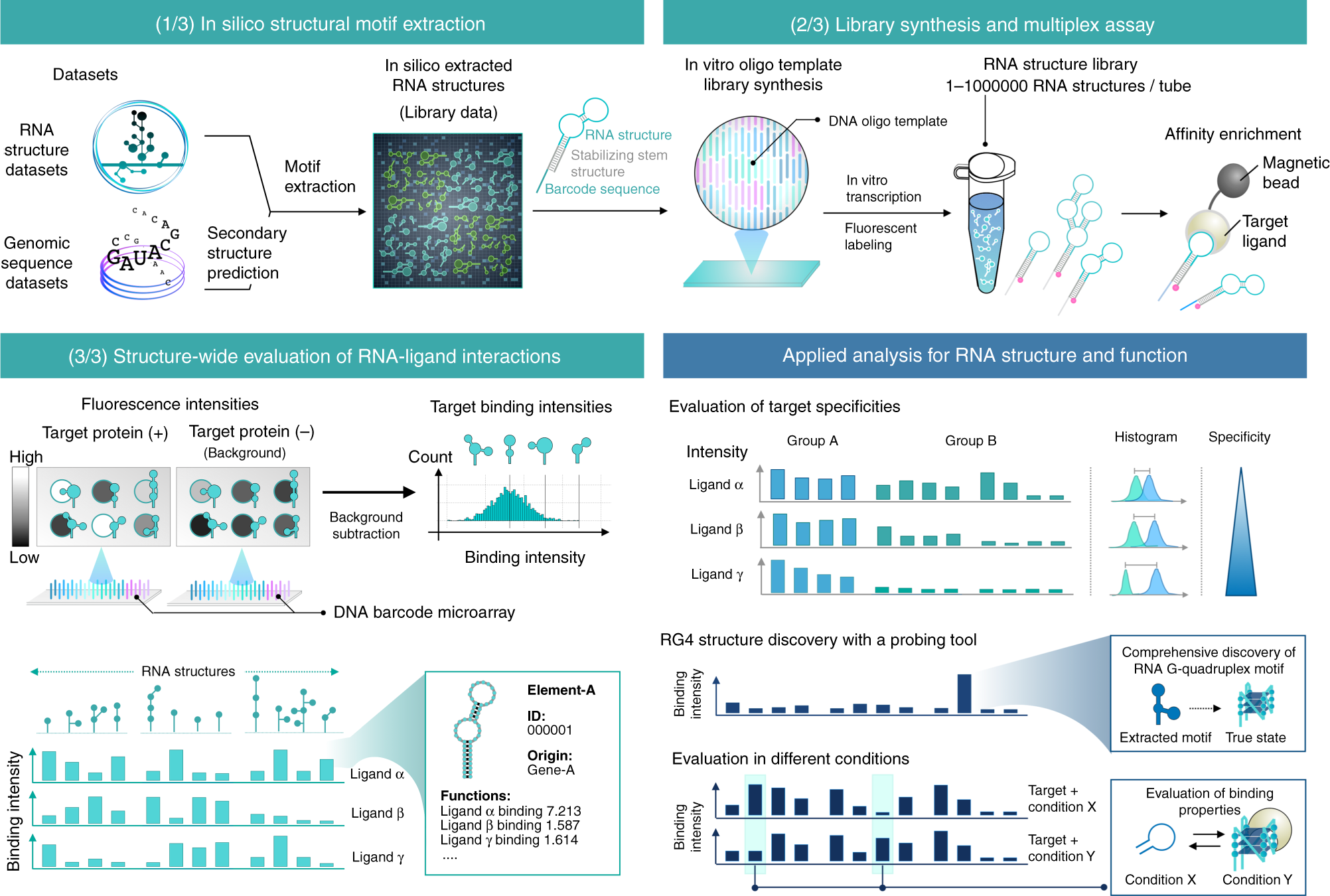
RNA structure-wide discovery of functional interactions with multiplexed RNA motif library | Nature Communications

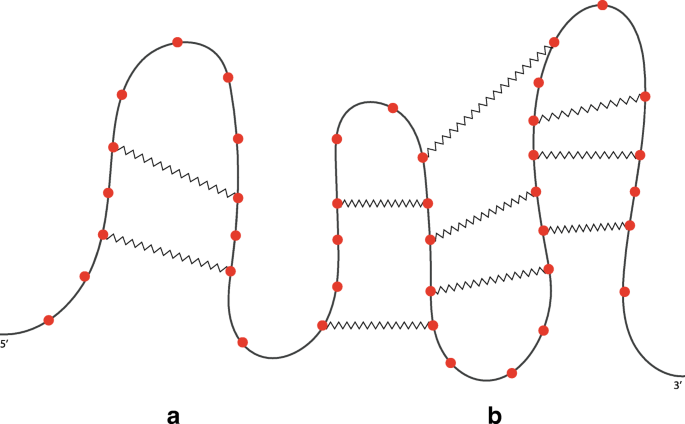
An RNA secondary structure illustrating the types of features included... | Download Scientific Diagram

Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms - ScienceDirect

Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: possible implications for −1 ribosomal frameshifting stimulation | Scientific Reports
RNA secondary structure prediction using deep learning with thermodynamic integration | Nature Communications

RNA structure types. (A) cartoon schematic of RNA structure types. (B)... | Download Scientific Diagram

Structural elements: the five structural elements of RNA structure,... | Download Scientific Diagram

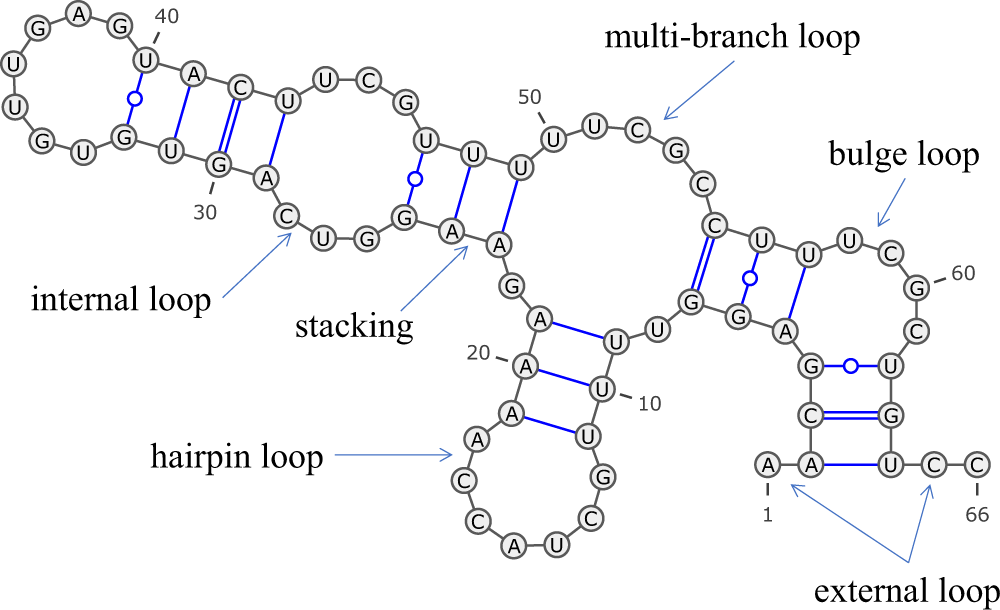
An RNA secondary structure with examples of the five kinds of loops:... | Download Scientific Diagram